A new DNA methylation regulator
CGGBP1 (CGG triplet repeat-binding protein 1) was first identified as a protein that binds unmethylated CGG repeats. It acts as a transcription regulator with target sites at CpG-rich sequences such as CGG repeats and Alu-SINEs (short interspersed elements) and L1-LINEs (long interspersed nuclear element). This protein is involved in several cellular processes like regulating cell proliferation, cell stress response, cytokinesis, telomeric integrity and transcription. However the role of CGGBP1 in the context of regulation of methylation on CpG regions has never been studied. Recently it has been shown that CGGBP1 binds to the transcription-regulatory regions of a subset of Alu-SINEs and L1-LINEs.

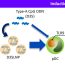
Fig. 1. (A) shows changes in CpG methylation. The increase in methylation is significant between CGGBP1-depleted and Control samples. Y-axis shows C count (%, [calculated as C count x100/total number of nucleotides]).
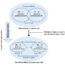
(B) is the frequency plotting of CpG methylation changes across different ranges of methylation. This plot shows binning of data depicted in 1A. X-axis shows methylation frequency bins and Y-axis shows C count (%).
By taking a whole genome and targeted methylation sequencing (at Alu and LINE-1 repeats) approaches we deciphered the role of CGGBP1 in regulation of CpG methylation. Our results show that upon depletion of CGGBP1 cytosine methylation increases (Fig. 1a). The exciting fact of this finding is that the increase in methylation was on genomic regions that were previously 70% – 90% methylated (Fig. 1b). Thus denoting that CGGBP1 depletion does not lead to anomalous methylation of unmethylated regions rather already methylated regions get slightly but significantly hyper methylated.
In case of Alu’s there was a bidirectional change in the methylation where major fraction has increase in methylation while a minor fraction has decrease of methylation while LINE-1 showed a significant increase after CGGBP1 depletion. Overall we have discovered a unique feature of CGGBP1 that is important for regulation of DNA methylation. This has implications on silencing of Alu and LINE-1 repeats, heterochromatin formation on simple and satellite repeats and hence on genome integrity and function.
Prasoon Agarwal
Department of Laboratory Medicine (LABMED),
Division of Clinical Immunology, Karolinska Universitetssjukhuset,
Stockholm, Sweden
Publication
CGGBP1 mitigates cytosine methylation at repetitive DNA sequences.
Agarwal P, Collier P, Fritz MH, Benes V, Wiklund HJ, Westermark B, Singh U
BMC Genomics. 2015 May 16











Leave a Reply
You must be logged in to post a comment.